Application of Deep Learning to Pancreatic Imaging – The Radiologists’ Perspective
Application of Deep Learning to Pancreatic Imaging – The Radiologists’ Perspective Linda C. Chu The Russel H. Morgan Department of Radiology and Radiological Science, The Department of Pathology, The Department of Cancer Research, and the Department of Computer Science, Johns Hopkins University, Baltimore |
Disclosure
|
Learning Objectives
|
Introduction
|
Deep Learning
|
|
Our Experience – The FELIX Project
|
Multidisciplinary Team Approach 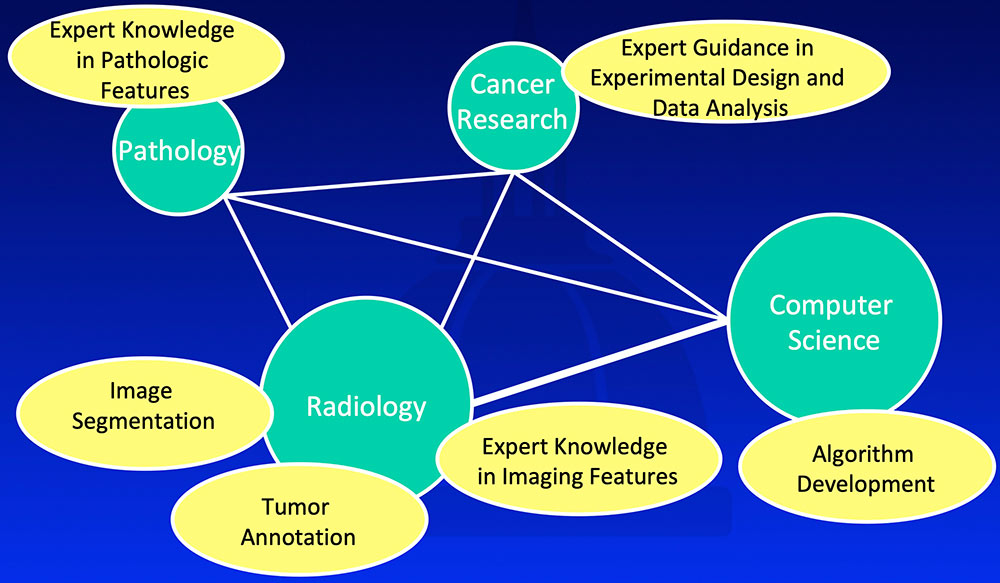 |
The FELIX Project
|
#1: Supervised Learning
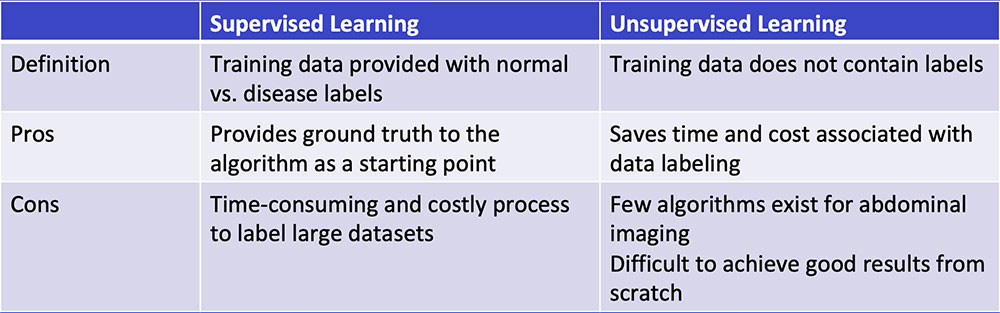 Chartrand G et al. RadioGraphics 2017;37:2113-2131. |
Overall Workflow 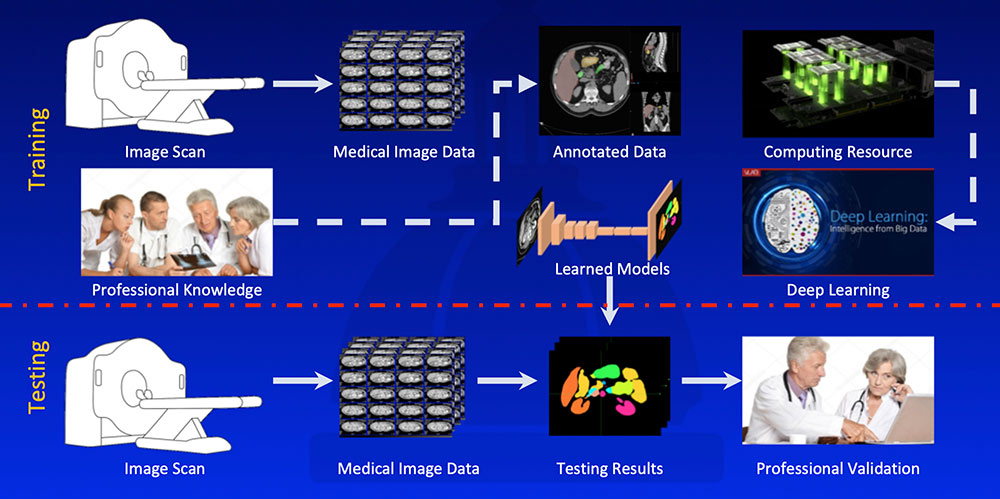 |
#2: High Quality Input Data
  Kawamoto S et al. [Submitted] |
#2: High Quality Input Data
|
#3: Learning Normal Anatomy
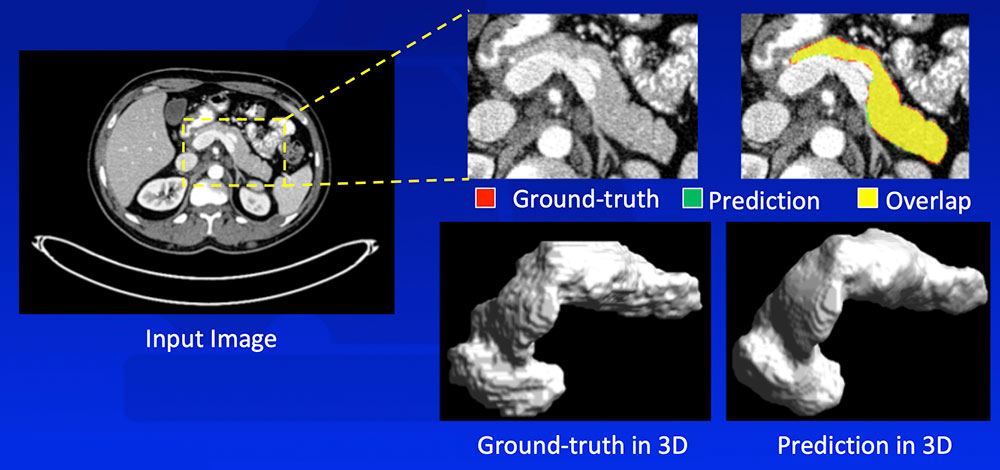 Zhou Y et al. arXiv:1612.08230. MICCAI, 2017. |
FELIX segmentation algorithm outperformed state-of-the-art algorithms on publicly available NIH dataset. It achieved even higher segmentation accuracy on the Johns Hopkins dataset, likely due to larger high quality dataset, thinner slices, and our experience. 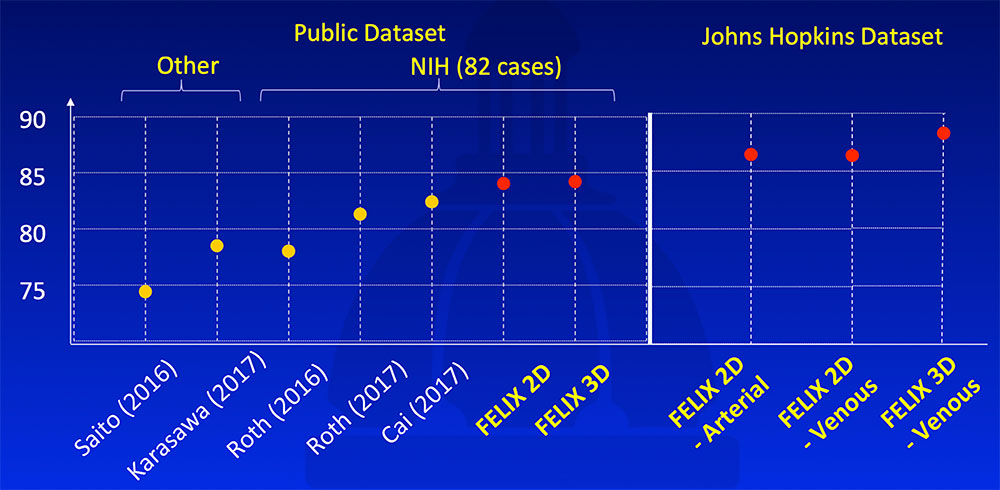 Saito A et al. Med Image Anal. 2016;28:46-65. Karasawa K et al. Med Image Anal. 2017;39:18-28. Roth H et al. arXiv:1606.07830. MICCAI, 2016. Roth H et al. arXiv:1702.00045. 2017. Cai J et al. arXiv:1707.04912. MICCAI, 2017. Yu Q et al. arXiv:1709.04518. CVPR, 2018. |
#3: Learning Normal Anatomy
|
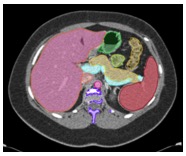
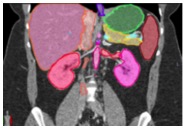
Multi-Organ Segmentation We have developed algorithms that can achieve >85% accuracy in segmentation of major abdominal organs 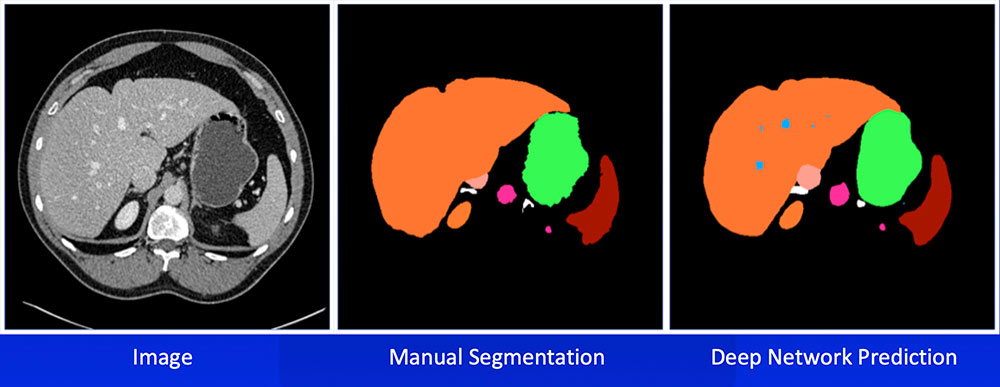 Wang Y et al. arXiv:1804.08414. |
Multi-Organ Segmentation Multi-organ segmentation (n = 575): 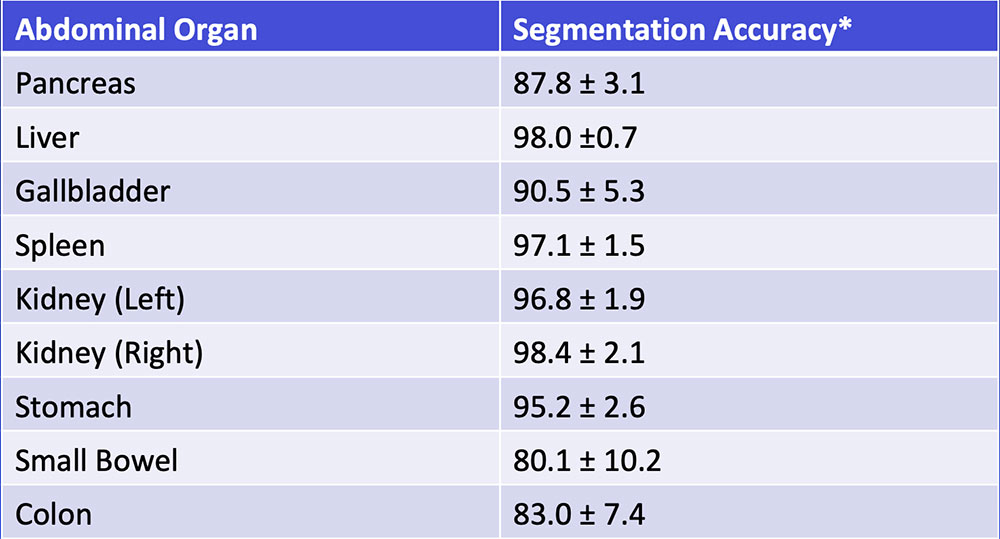 Wang Y et al. arXiv:1804.08414. Kawamoto S et al. [Submitted] |
Multi-Organ Segmentation
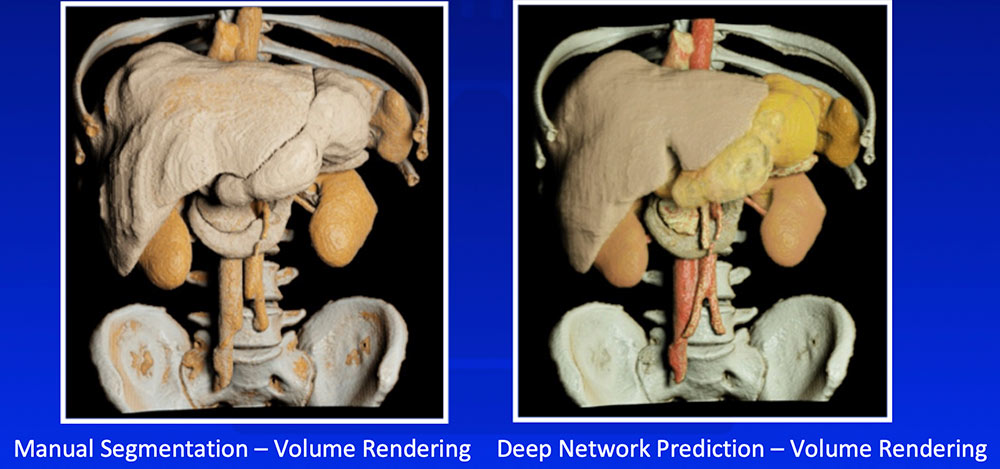 Wang Y et al. arXiv:1804.08414. |
#4: Recognizing Tumor
|
#4: Recognizing Tumor We have developed a number of deep learning algorithms that can recognize pancreatic ductal adenocarcinoma based on abnormal shape and/or texture 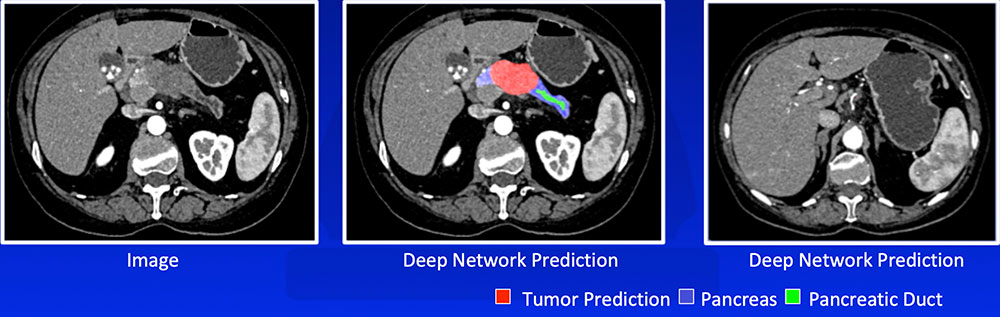 Liu F et al. arXiv:1804.10684. Zhu Z et al. arXiv:1807.02941. |
#4: Recognizing Tumor
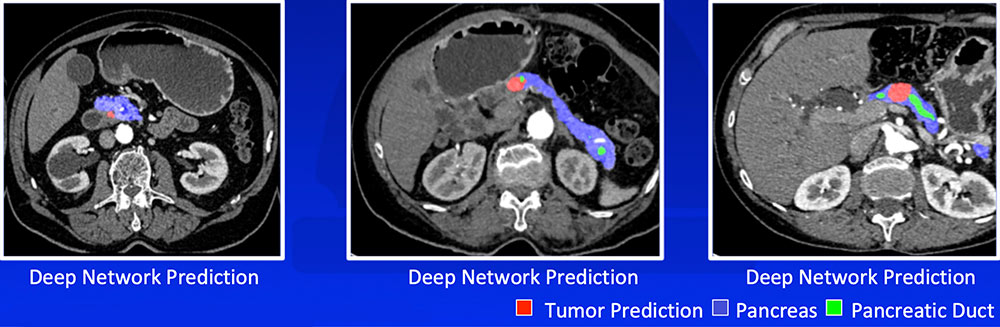 Liu F et al. arXiv:1804.10684. Zhu Z et al. arXiv:1807.02941 |
#5: Troubleshooting
|
#5: Troubleshooting 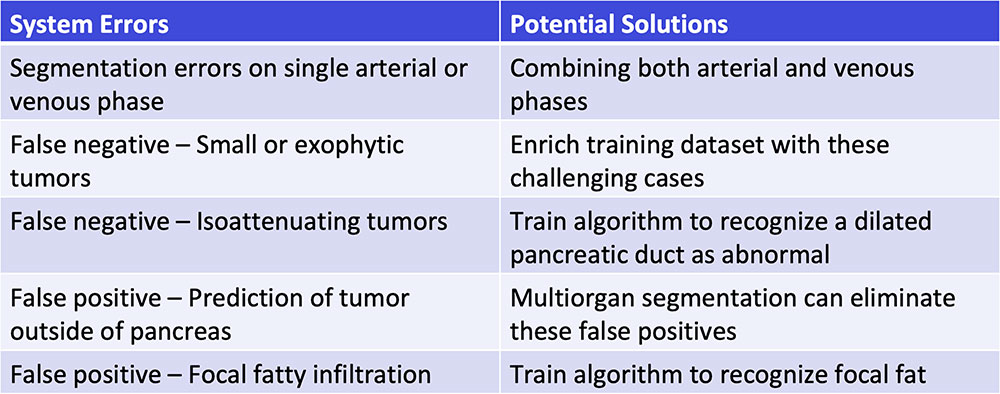 |
Single-Phase vs. Dual-Phase
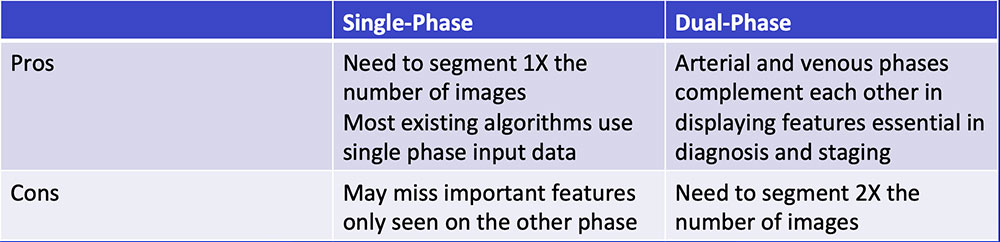 |
Single-Phase vs. Dual-Phase Combining arterial and venous phase can improve segmentation accuracy of the pancreatic tumor by improving segmentation accuracy of adjacent organs and vasculature 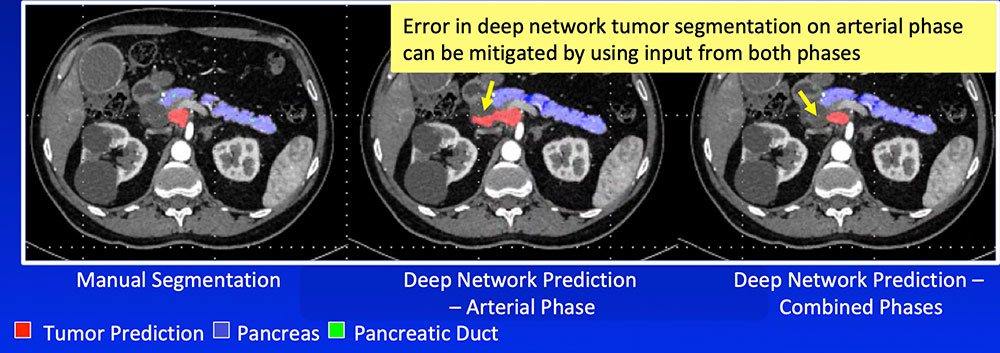 |
False Negative – Small or Exophytic Tumors
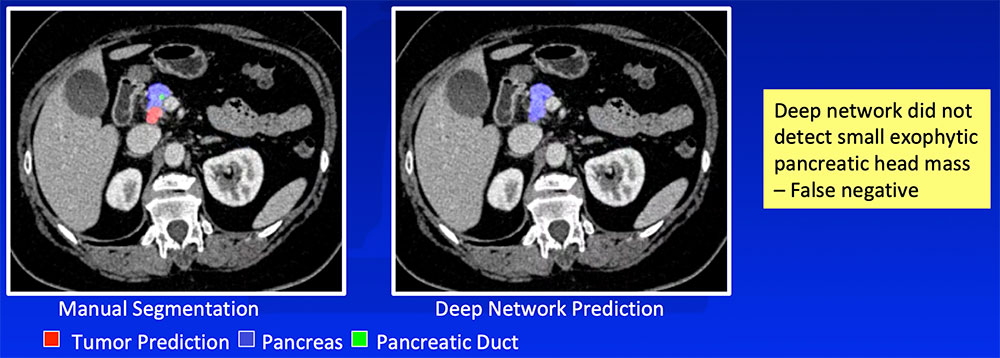 |
False Negative – Small or Exophytic Tumors
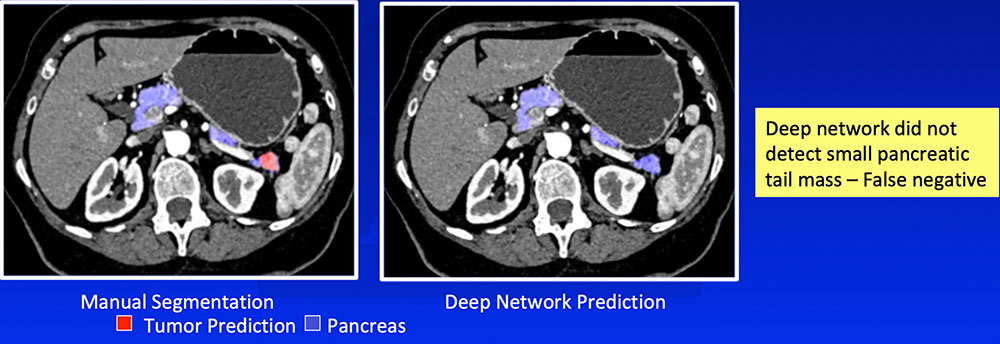 |
False Negative – Isoattenuating Tumors
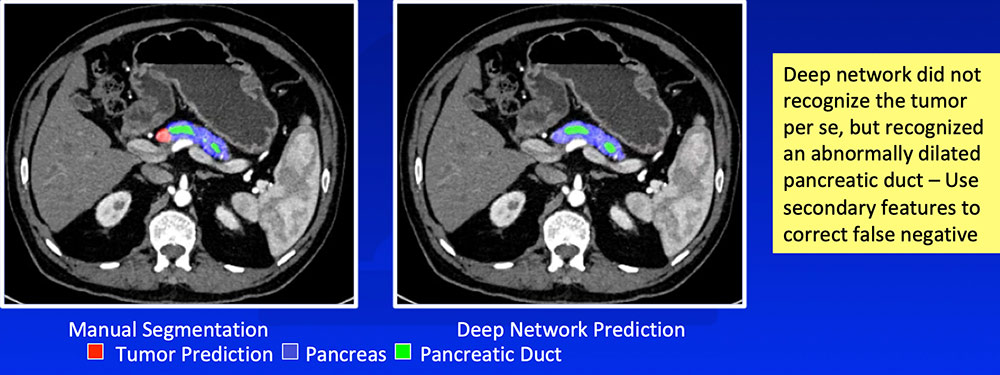 |
False Positive – Prediction of Tumor Outside Pancreas Application of multi-organ segmentation algorithm can correct for false positives where the predicted tumor is located at the predicted location of another organ 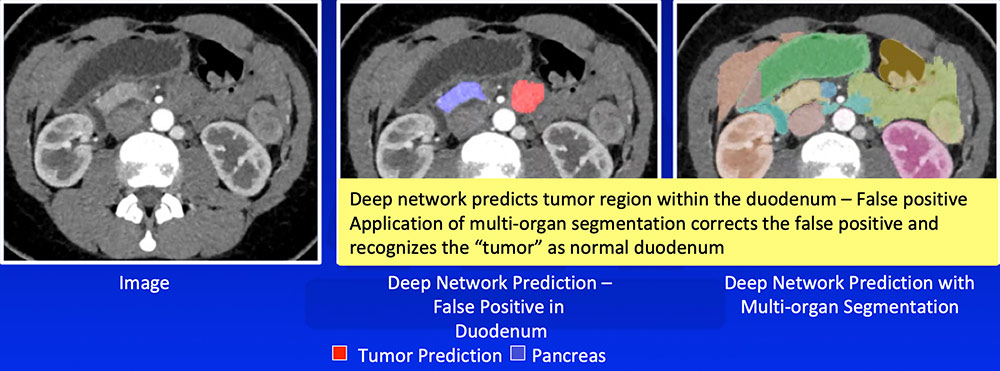 |
False Positive – Focal Fat
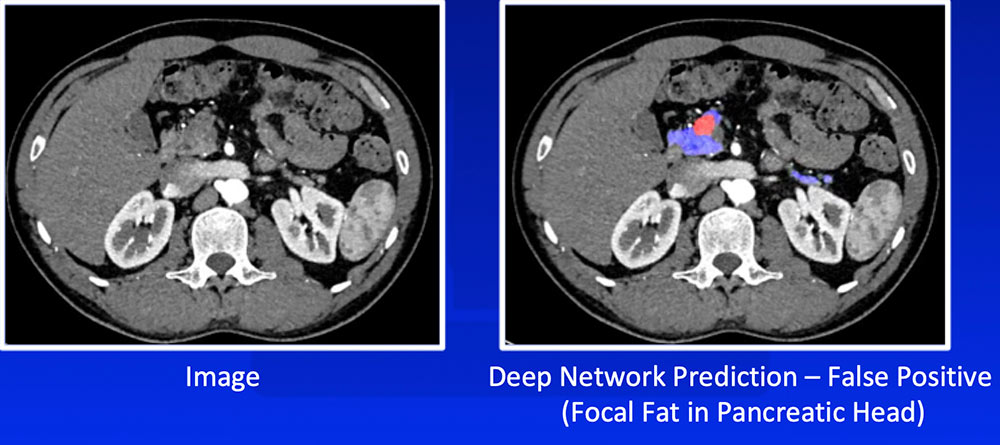 |
Future Directions
|
Future Directions
|
Conclusion
|
References
Ackownledgements
|
